Navigation auf uzh.ch
Navigation auf uzh.ch

We study the properties of disordered and heterogeneous systems out of equilibrium. This encompasses light transport in turbid media and photonic glasses, with applications in imaging, structural colours, light harvesting for energy and secure optical communication. A second focus of our activities is the study of the elastic properties of growing biological tissues and their influence on development and pattern formation, e.g. in the regeneration of zebrafish fins or the process of dorsal closure in drosophila embryos. In all these fields our investigations are mainly experimental, developing the tools necessary, e.g. to study forces in tissues on the scale from nN to mN, however we also use computational modeling to guide these experiments.
Modelling Dorsal Closure in Drosophila embryos
Tissue morphogenesis integrates cell type-specific biochemistry and architecture, cellular force generation and mechanisms to coordinate these forces amongst neighbouring cells and tissues. Here, we use finite element-based modelling to explore the interconnections at these multiple biological scales in the developmental process of dorsal closure, where pulsed actomyosin contractility in adjacent Amnioserosa (AS) cells powers the closure of an epidermis opening. We cross the different scales by modelling the biochemical regulations as well as the visco-elastic tissue properties in the same finite element framework, with cells distributions based on direct observations in vivo. Furthermore, we base the input of our biochemical regulation model on in vivo observations by our collaborators, for instance implementing F-actin nucleation periodicity that is independent of MyoII activity. Given these few and well established regulatory mechanisms, our model can reproduce several emergent properties of the dorsal closure process, such as the tissue closure or the arrest of cell pulsations. This questions the previously proposed role of Dpp-mediated regulation of the patterned actomyosin dynamics in the AS tissue, suggesting them to be emergent. Moreover, the model also shows how depleting MyoII activity can under certain conditions nevertheless indirectly affect oscillatory F-actin behaviour without the need for biochemical feedback.
Figure: Snapshots of the simulation at different times showing both the contraction of the AS tissue as well as the increase in F-actin concentration during the process of dorsal closure.
The model further predicts that the mechanical properties of the surrounding epidermis tissue feed back on the shaping and patterning of the AS tissue again showing, how multiple scales interact to produce the observed final process. Finally, our model’s parameter space can reproduce mutant phenotypes and provides predictions for their underlying cause due to the identification of the different parameters with biological processes. Our modelling approach thus reveals several unappreciated mechanistic properties of tissue morphogenesis and lays the ground for an experimental investigation of these identified causes by developmental biologists.
Highlighted Publications:
1. In-vivo force measurements of MyosinII waves the yolk surface during Drosophila dorsal closure, L.Selvaggi et al.,Biophysical Journal 121 (2022), https://doi.org/10.1016/j.bpj.2021.12.038
2. Mechanochemical modelling of dorsal closure emergent cell behaviour and tissue morphogenesis, F. Atzeni et al., bioRxiv 2020.01.20.912725, https://doi.org/10.1101/2020.01.20.912725
3. Stochastic Resonance in Noisy Optical Resonators Modeled Through Coupled-Mode Theory, V. Mazzone and C.M. Aegerter, Communications in Nonlinear Science and Numerical Simulation (2022), http://dx.doi.org/10.2139/ssrn.4312925
We are conducting research and development in Medical Physics, Theoretical Biology and Medical Modelling. Our main topics are: Development of radio-biological models, space radiation research, Monte Carlo simulations and dosimetry for radiotherapy and imaging and the development of novel detector systems.
Tumor control probability (TPC) models based on Poisson statistics characterize the distribution of surviving clonogens, thus enabling the calculation of the TCP for individuals. To mathematically describe clinically observed survival data of patient cohorts it is necessary to extend the model. This is typically done by using an empirical logistic model. We developed an analytical population TCP model by establishing a mechanistic link between the Poisson statistics based individual and the logistic TCP model. This link can be used to determine the radiobiological parameters of patient specific TCP models from published fits of logistic models to cohorts of patients.
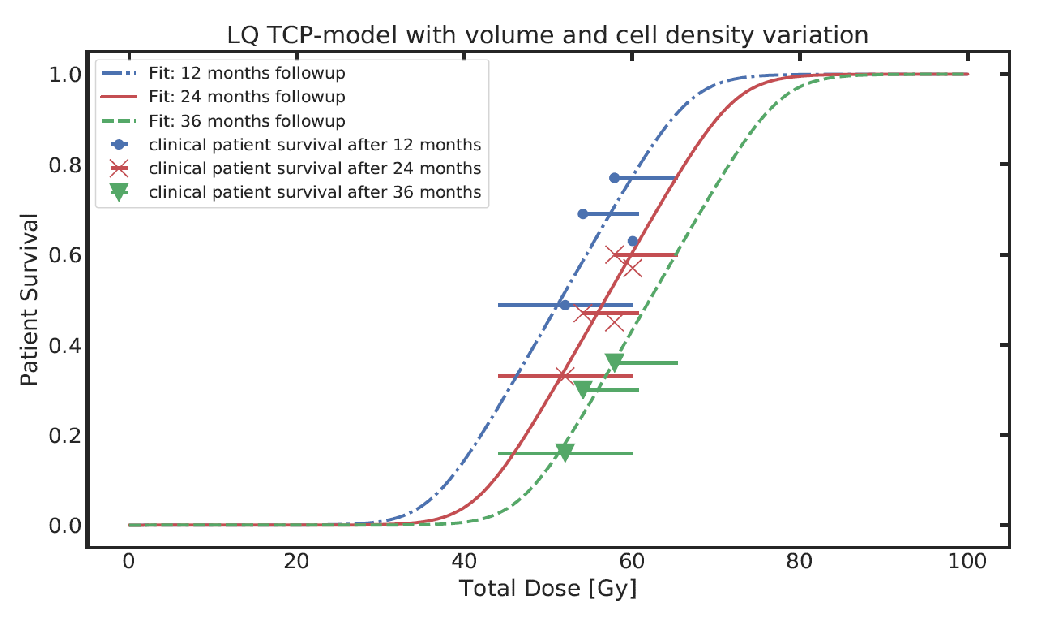
Figure: Fitting clinical patient survival with extended TCP population model incorporating tumour volume and cell density variations within a patient population.
Highlighted Publications:
Tumour volume distribution can yield information on … U. Schneider, J. Besserer et al., Z Med Phys. 2022 Jun 9:S0939-3889(21)00053-2
Radiotherapy is one of the mainstays of cancer treatment and a highly technology-driven field of medicine. Our research group contributes to the development of radiotherapy technology by applying concepts from physics, mathematics, statistics, and machine learning to problems in medical imaging and radiation oncology.
We focus on three areas of research:
1) Radiotherapy treatment planning: We work on mathematical optimization methods to optimally combine x-ray and proton beams, and to optimally distribute radiation dose.
2) Target delineation and outcome prediction: Here, we focus on quantitative modeling of tumor progression and the analysis of medical images such as MRI, CT, and PET, with the goal of precisely defining the region to be irradiated and predicting the patient’s response to treatment.
3) Adaptive radiotherapy: The MR-Linac, a combination of MRI scanner and radiotherapy device allows MR imaging of a patient during treatment. We work on innovative concepts to use the technology, e.g. MR-only workflows [1] and the concept of adaptive fractionation [2].
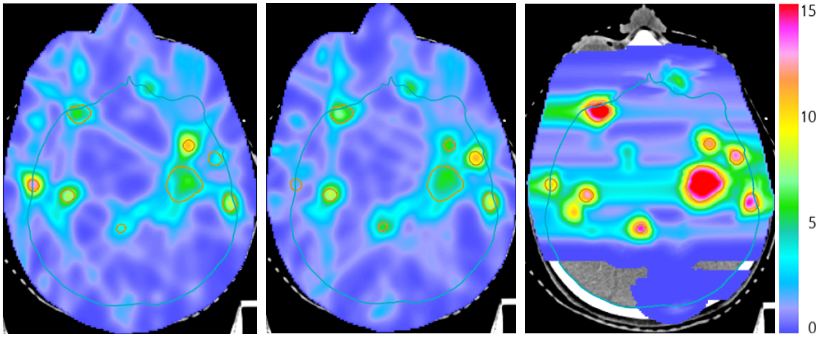
Figure: Combined proton-photon radiotherapy for a patient with many brain metastases treated with one proton fraction (right panel) and two photon fractions. The superior proton dose distribution is exploited by delivering larger doses.
Highlighted Publications:
1. M. Lapaeva et al., Phys Imaging Radiat Oncol. 2022 Nov 28;24:173-179
2. Y. Pérez Haas et al. Phys. Med. Biol., 19;68(3), 2023
We study the structure, dynamics, and functions of biomolecules, especially proteins, the nanomachines of life. Towards this goal, we develop and apply single- molecule fluorescence and force spectroscopy, often in close combination with theory and simulations. A particularly important tool is Förster resonance energy transfer (FRET), a spectroscopic nanoscale ruler.
We recently started employing nanophotonics to enhance the fluorescence emission of single molecules, which allows us to probe previously inaccessible nanosecond motion in proteins and directly compare to molecular dynamics simulations [1] (see Figure). An interesting discovery was a new molecular mechanism of protein interactions that is important for regulating DNA [2].
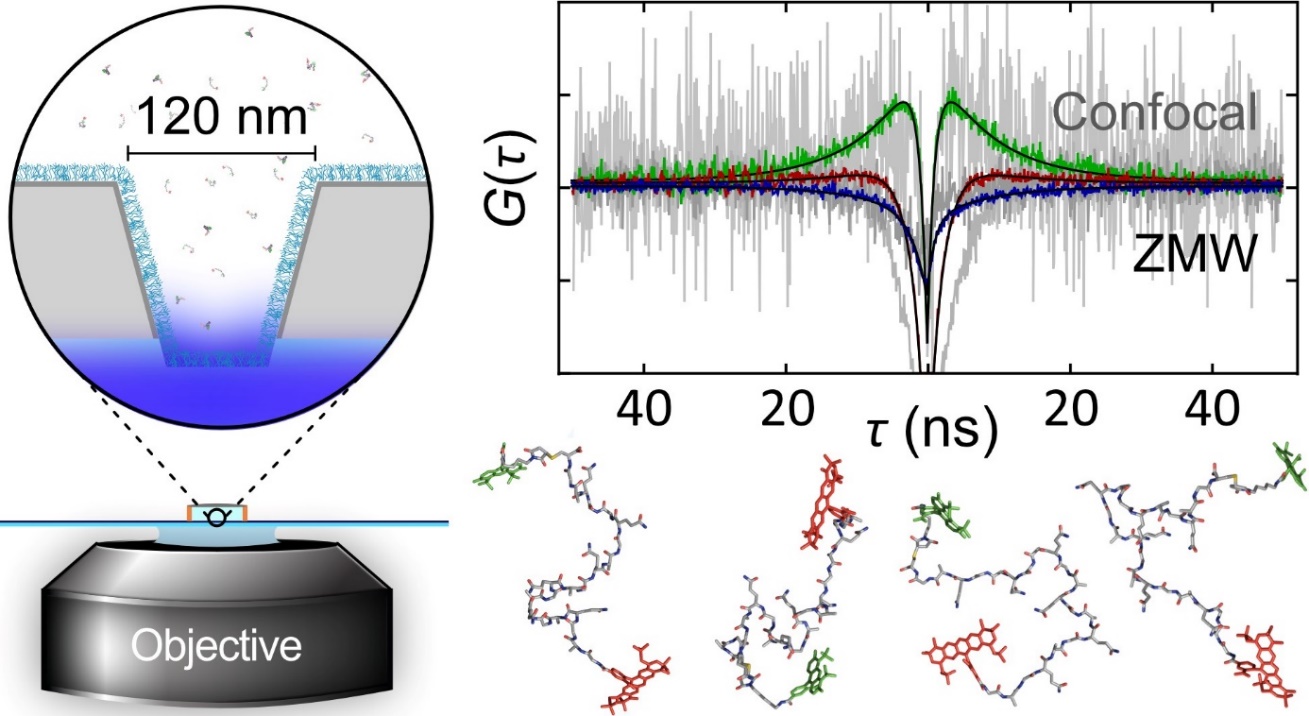
Figure: Zero-mode waveguides nanofabricated in thin aluminium layers enhance fluorescence emission to a level that ultrafast motions within proteins become detectable with nanosecond correlation spectroscopy. Snapshots from computer simulations illustrate some of the myriad molecular configurations.
Highlighted Publications:
1. Single-molecule detection of ultrafast biomol. … M. F. Nüesch et al., J. Am. Chem. Soc. 144, 52-56
2. Release of linker histone from the nucleosome … P. O. Heidarsson et al., Nat. Chem. 14, 224-231